



Welcome to the Gupta Lab
We are fascinated by the differential regulation of microtubule dynamics within cells. Microtubules are cytoskeletal filaments of polymerized tubulin that are required for many cellular processes including cell division, cell migration, and nervous system development. A critical but poorly understood feature of microtubules, which enables such diverse activities, is that their assembly and disassembly are regulated in space and time by a wide range of interacting proteins. The importance of regulating microtubule dynamics is highlighted by two significant impacts on human health – mutations in tubulin result in human neurological disorders and inhibiting microtubule dynamics is a proven anti-cancer therapy. The Gupta lab is focused on determining the mechanisms that control microtubule dynamics in healthy cells, and using our expertise in tubulin biochemistry to understand how defects in microtubule dynamics cause neurological disease and can be leveraged for improved cancer treatment. To learn more about our research please click on the 'research areas' tab on the menu above.
Differential Regulation of Microtubule Dynamics and Function
Microtubules are essential cytoskeletal filaments of polymerized tubulin protein. Microtubules can undergo dynamic cycles of polymerization and depolymerization within the cell. This dynamic behavior of microtubules underlies numerous, critical processes throughout biology including organism development, neurogenesis, intracellular transport and cell division. To understand these highly complex and transient events, we must define how microtubule dynamics are controlled across multiple scales. We use a combination of genetics, cell biology, quantitative live cell imaging, in vitro reconstitution and single-molecule TIRF microscopy. With this ‘cellular biophysics’ approach we study microtubule function at the scale of single tubulin molecules, the scale of the microtubule polymer, and the integration of microtubule dynamics with spatial cues and cellular processes. We investigate temporal scales ranging from the dynamic behavior of individual filaments to the higher order organization needed for chromosome segregation and spindle positioning across the cell cycle.
Microtubules and their regulatory proteins are highly conserved. Thus, although we work with metazoan systems when appropriate, we exploit the incomparable experimental tools available in the budding yeast model, S. cerevisiae. Higher organisms express multiple isoforms of tubulin, and obtaining biochemical amounts of recombinant mammalian tubulin remains a challenge. The fact that yeast is viable with only a single isoform, and the ability for direct gene replacement make it the leading system for tubulin structure-function analysis. Another striking advantage of yeast is that we can visualize the dynamic behavior of individual microtubules performing discrete tasks.
Please see the following sections for a summary of our efforts to determine how microtubule dynamics are influenced by the properties of the tubulin molecule, regulated at the level of the microtubule polymer, and integrated into diverse cellular processes. Our overall goal is to reveal key insights required to understand how microtubule dynamics are regulated for normal growth and development, and to exploit these mechanisms for improved human health.
Kinesin motor proteins power movement along microtubules at the molecular level, but they also play important roles in controlling microtubule dynamics. Our work discovered that the highly conserved Kinesin-8 is a multifunctional motor that combines motility with the ability to both destabilize and stabilize microtubules (Gupta et al., 2006, Nature Cell Biology, pdf; Su et al., 2011, Molecular Cell, pdf). Kinesin-8 is required for the cells of higher eukaryotes to survive. This makes it difficult to study the effects of Kinesin-8 loss in more complex organisms. Thus, determining how Kinesin-8 regulates microtubule dynamics in the simplified and tractable yeast cell is an ideal model for understanding their biological roles in higher eukaryotes, including human cells.
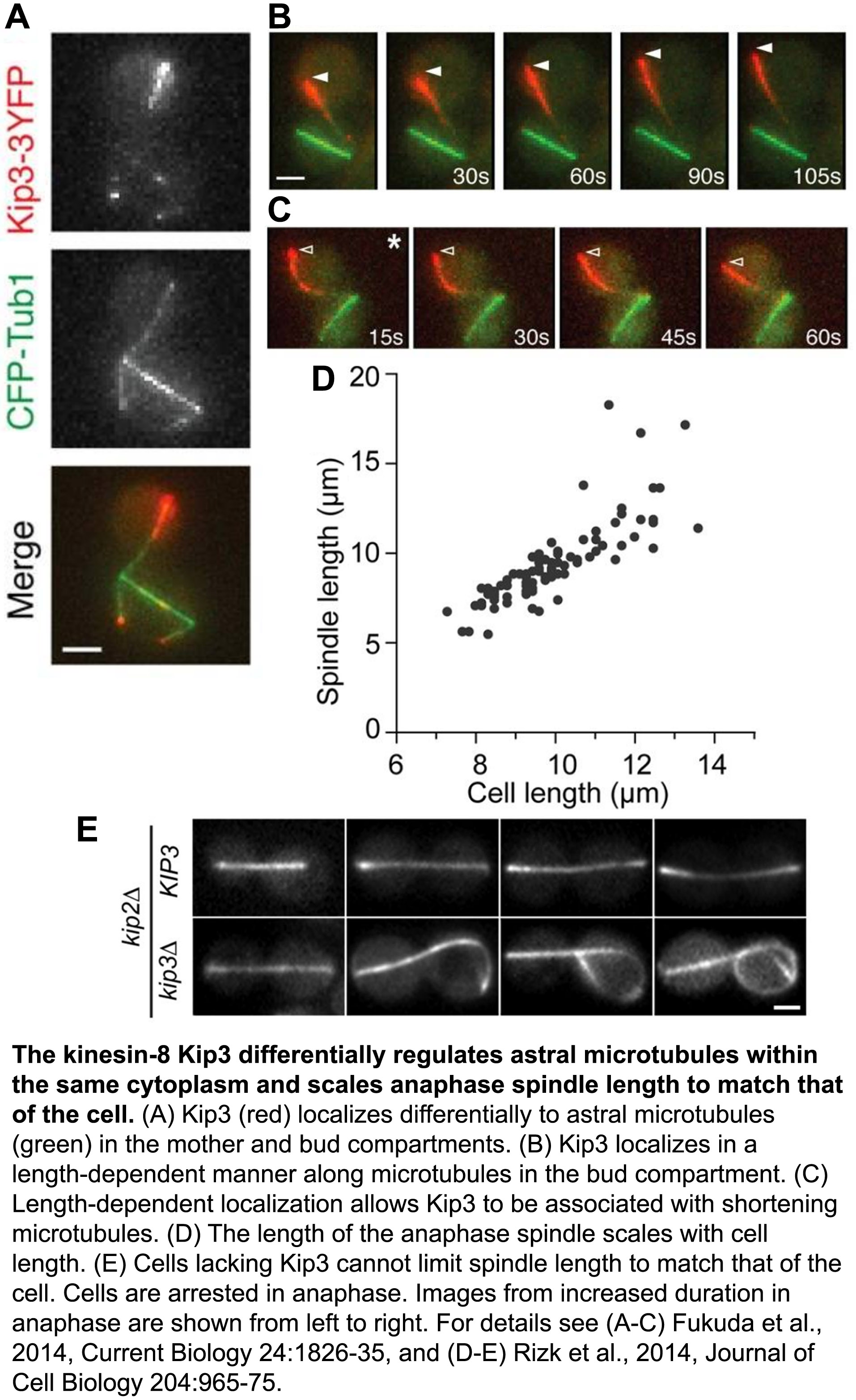
The ability of cells to differentially control the length or longevity of individual microtubules within the shared cytoplasm of a single cell is largely unknown. Focusing on astral microtubules during the process of spindle positioning, we elucidated how distinct Kinesin-8 activities are deployed spatially within the cell to stabilize or destabilize specific microtubules (Fukuda et al., 2014, Current Biology, pdf). We found that the destabilizing activity of Kinesin-8 is enhanced when microtubules reach one end of the cell, which appropriately tunes their length to that of the cell (Fig. 1). We also discovered a novel mechanism by which Kinesin-8 associates with, and selectively stabilizes a subset of shortening microtubules to dramatically modulate their lifetime. Physiologically, we revealed how this differential control of microtubule stability helps prevent mitotic exit when spindles are mispositioned, thus maintaining genome stability.
The regulated morphogenesis and bipolar structure of the microtubule-based mitotic spindle are essential for cell division. Spindle elongation during anaphase ensures chromosome segregation. Yet, to maintain structural integrity of the bipolar spindle, spindle length cannot exceed cell diameter. Control of anaphase spindle length has been observed in diverse species, but the underlying mechanisms remain obscure. We discovered that the budding yeast Kinesin-8, Kip3, is the key regulator of spindle length during anaphase (Rizk et al., 2014, Journal of Cell Biology, pdf). In particular, our work revealed how Kinesin-8’s depolymerase activity opposes the action of microtubule polymerases spatially within the spindle to control elongation forces and scale anaphase spindle length to match cell diameter.
Going forward we are focused on how cells control the dynamics of specific microtubules both spatially and temporally within a single cell. We will continue to build on our knowledge by leveraging the strengths of the yeast model system including live cell imaging, molecular genetics, and synchronization of cell cultures precisely when Kinesin-8 is regulating specific classes of astral microtubules. Our results will define mechanisms that allow cells to differentially control individual microtubules within a common cytoplasm. Additionally, we seek to understand how diverse regulatory proteins cooperate with Kinesin-8 to allow the morphogenesis, yet ensure the structural integrity of the microtubule cytoskeleton.
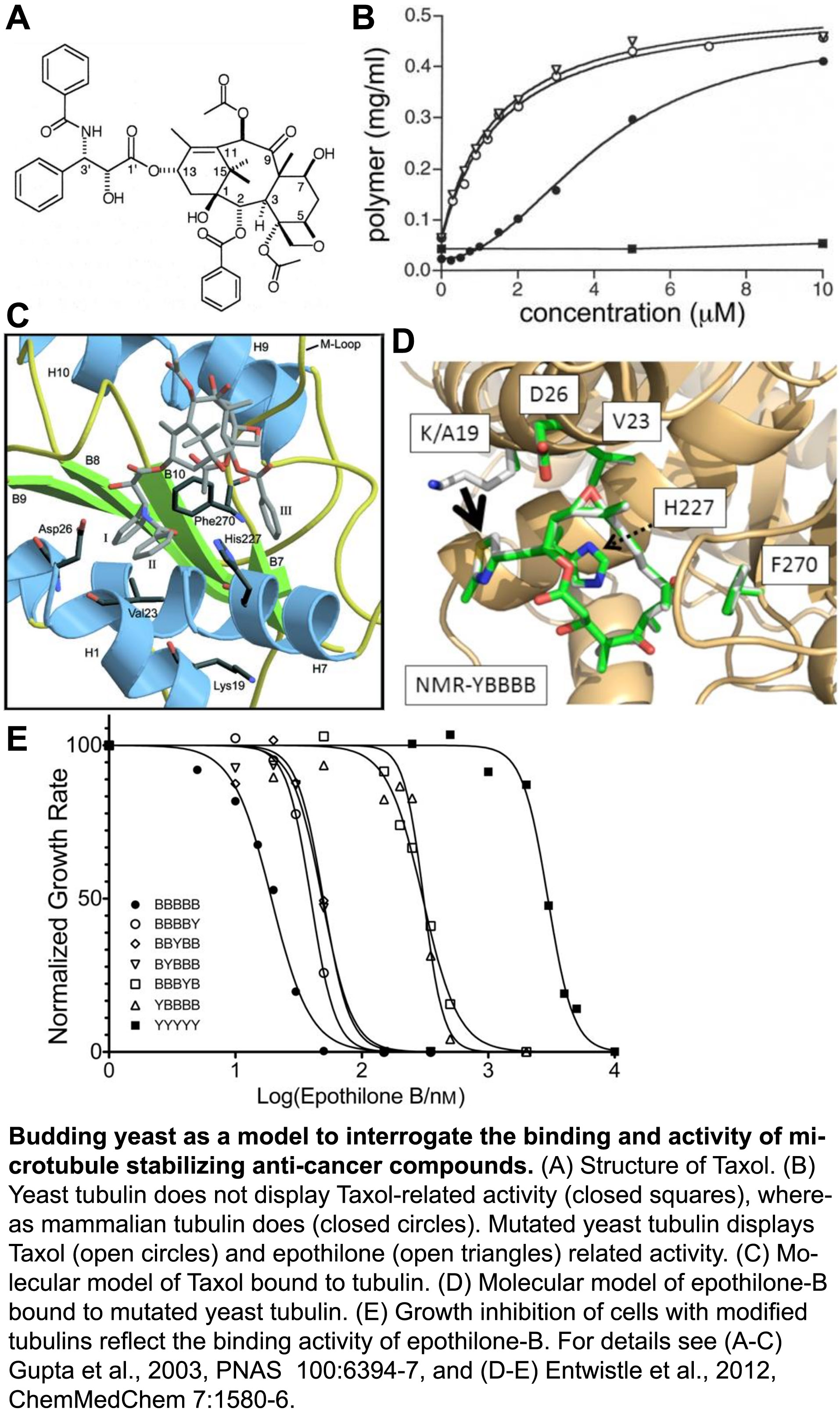
To ensure proper chromosome segregation during cell division, sister chromatids must establish attachments with microtubules from opposite spindle poles. Only in this bipolar configuration can the dynamic microtubules generate tension across the sister kinetochores. In cells, a surveillance mechanism called the Spindle Assembly Checkpoint (SAC) prevents anaphase until chromatids are properly attached, and SAC failure results in chromosome missegregation, which can lead to birth defects and more aggressive tumors. However, microtubule-stabilizing compounds like Taxol are powerful cancer treatments because they disrupt prevent proper kinetochore attachments, which activates the SAC and induces cell death.
Despite a central role in guarding genome stability, whether the SAC is triggered by defects in microtubule attachment to kinetochores, insufficient tension on kinetochores, or both. A major limitation has been the experimental intractability of tension and attachment in current experimental systems. Briefly, treatments that inhibit kinetochore attachment also preclude MT-generated tension. Thus, reduced tension cannot be excluded as a SAC signal. Conversely, methods to reduce tension have been shown to induce kinetochore detachment. In higher eukaryotes, MT stabilizers such as Taxol reduce tension, but their kinetochores bind many MTs and Taxol reduces the number bound, causing ‘partial detachment’ which, again, cannot be excluded as a SAC signal. Notably, yeast kinetochores bind just one microtubule each, so they can’t suffer partial detachment. However, Taxol does not bind to yeast microtubules. Thus, tension and attachment are difficult, if not impossible, to experimentally separate in normal cell systems.
We developed a unique experimental system that allows the question of tension versus attachment to be squarely addressed. To investigate the interactions between Taxol-like compounds and microtubules, we engineered an active Taxol-binding site into yeast tubulin (Entwistle et al., 2012, ChemMedChem, pdf). We have now developed this Taxol-sensitive yeast as a system to reduce tension at otherwise fully attached kinetochores. With this system in hand, we are actively investigating how the SAC responds to reduced tension at kinetochores, independent of the attachment status.
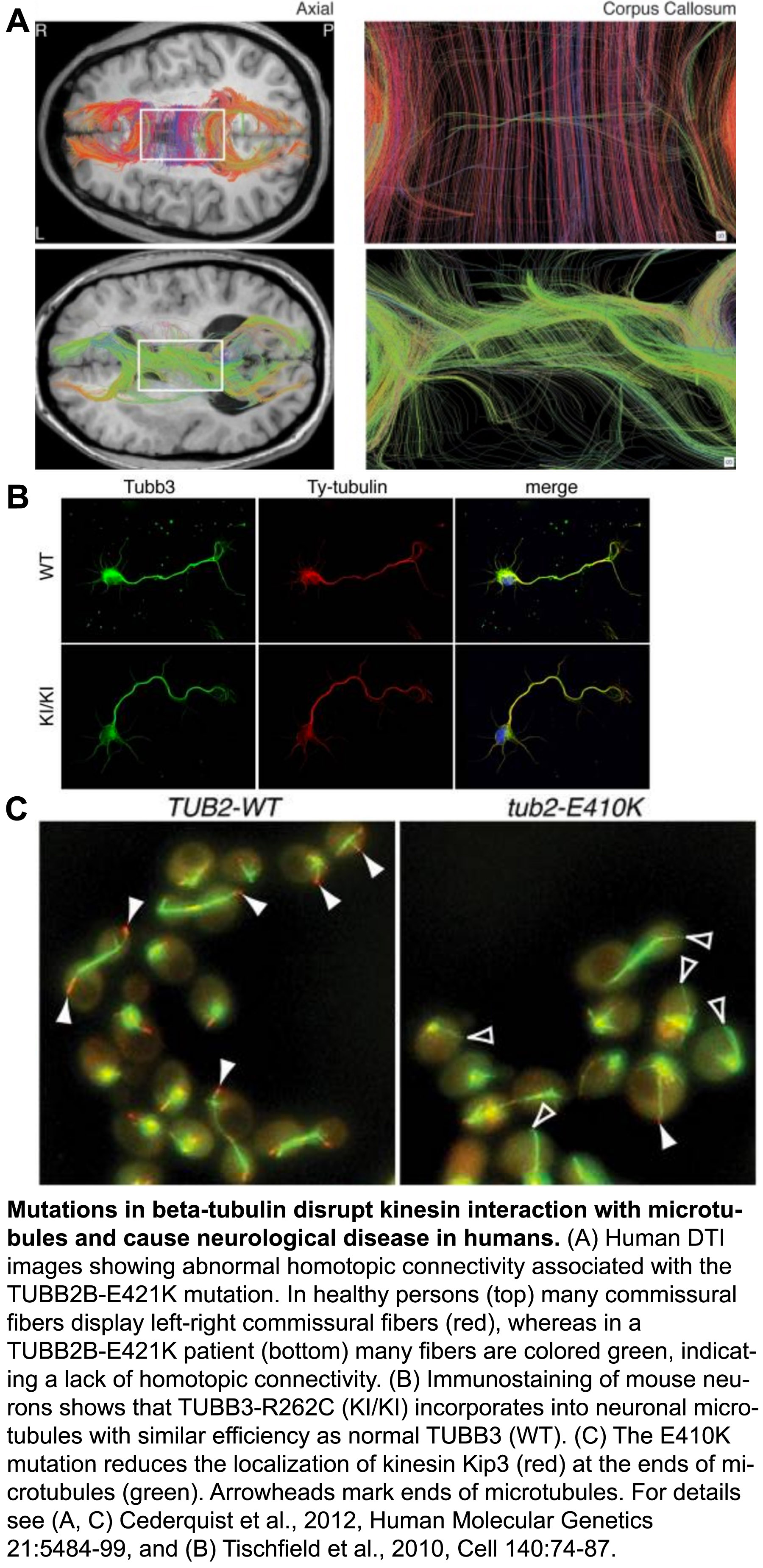
The tubulin protein is a heterodimer consisting of an α- and a β-subunit, and mammals have ~eight versions, or isoforms of each. These isoforms have specialized, but poorly understood functions in different types of cells. Over the last few years, it’s become clear that a range of mutations in various isoforms result in the ‘tubulinopathies’, a spectrum of human disorders characterized by abnormal neuronal migration and defects in axon guidance/maintenance.
We have utilized our expertise with tubulin structure-function analysis to elucidate the mechanisms underlying the tubulinopathies. In a series of studies we discovered how distinct mutations in the β-isoforms TUBB3 and TUBB2B, which cause related axon disorders, dominantly increase microtubule stability and disrupt kinesin function. Together with our collaborators, we revealed the genotype-phenotype relationship at a depth not possible in mammalian models, which confirmed these properties likely underlie disease progression (Tischfield et al., 2010, Cell, pdf; Cederquist et al., 2012, Human Molecular Genetics, pdf). In another study we demonstrated how various mutations in the primate specific β-tubulin isoform, TUBB8, cause human infertility by dominantly disrupting microtubule dynamics and function (Feng et al., 2016, New England Journal of Medicine, go to journal article. Our paradigm-building work established that the tubulinopathies result from diverse, yet dominant mutations, and that the distinct disorders may result from specific changes to the mutant microtubule properties. Moving forward we will continue to leverage the strengths of the yeast system to understand how mutations of various isoforms affect microtubule dynamics and their ability to execute specialized functions. Our goals are to understand the molecular basis of tubulin-based diseases, and to uncover the physiological roles of individual tubulin isoforms.
Current Members

Graduate Student
kofiaa at iastate.edu

Undergraduate Student
""

Undergraduate Student
""

Principal Investigator
mgupta at iastate.edu

Undergraduate Student
""

Undergraduate Student
""

Undergraduate Student
""

Graduate Student
maliha at iastate.edu

Undergraduate Student
""

Undergraduate Student
""

Undergraduate Student
""

Undergraduate Student
""

Undergraduate Student
""

Undergraduate Student
""

Undergraduate Student
Former Members
Samuel Anderson, M.S.
Research Associate
Hemal Amin
Research Associate
Advanced to Research Scientist with BD Biosciences, NJ.
Mitch Andrews
Undergraduate Student
Claire Baumer
Undergraduate Student
Advanced to Biology Graduate Program, Stanford University.
Abesh Bera
Graduate Student
Advanced to Immunochemistry Division, Elanco Animal Heath.
Angela Bunning
Graduate Student
Advanced to R&D Scientist at Luminex Corporation.
Daniel Cheng
Undergraduate Student
Ellie Clark
Undergraduate Student
Advanced to Biology Graduate Program, KU Leuven University.
Sandeep Dave
Postdoctoral Fellow
Sandra Orellana Diaz
Undergraduate Student
Katie DiScipio
Undergraduate Student and Research Associate
Advanced to MSTP Program, University of Connecticut School of Medicine.
Grace Hagedorn
Undergraduate Student
Advanced to Research Associate, University of Wisconsin - Madison, School of Medicine.
Joshua Hendin
Research Associate
Advanced to Stritch School of Medicine, Loyola University Medical Center.
Yusuke Fukuda
Graduate Student
Advanced to Postdoctoral Fellowship, Dana-Farber Cancer Institute/Harvard Medical School.
Cassandra Iroz
REU Undergraduate Student, Carleton College
Advanced to Graduate Program in Health Communication, Northwestern University.
Allison Juntunen
Undergraduate Student
Advanced to Graduate School of Public Health, Boston University.
Arvin Kannoly
Undergraduate Student
Seo Young Kim
Undergraduate Student
Advanced to Graduate Program, University of Texas - Austin.
Paulina Leduchowska
International Exchange Graduate Student
Ania Luchniak
Graduate Student
Advanced to Postdoctoral Fellowship, Yale University.
Joey Marcuccilli
Undergraduate Student
Erin Murphy
Research Associate
Advanced to Molecular Biologist, ACGT, Inc., Wheeling, IL.
Mohammad Bin Naveed
Undergraduate Student
Grant Nickles
Undergraduate Student
Emmanuel Nsamba
Graduate Student
Advanced to Postdoctoral Fellowship, Stanford University.
Ruchee Patel
Undergraduate Student
Rachel Plumb
REU Undergraduate Student, Oberlin College
Advanced to Yale Graduate Program in Cell Biology.
Kate Proudfoot
Graduate Student
Advanced to Science writer / account executive at CG Life.
Andrew Rapoport
Undergraduate Student
Rania Rizk
Postdoctoral Fellow
Advanced to Author and Senior Lecturer, RC Medreview, Roanoke, VA.
Karlas Delano Robinzine
Undergraduate Student
Sunil Rohatgi
Undergraduate Student
Advanced to the College of Medicine, University of Florida.
Pallavi Sinha Roy
Graduate Student
Advanced to Target Discovery Scientist with Loxo Oncology at Lilly.
Landon Savoy
Graduate Student
Vaishali Todi
Graduate Student
Advanced to Research Scientist with Elanco Animal Heath.
Shandra Truong
Undergraduate Student
Advanced to Research Associate at University of Iowa.
Slyn Uaroon
Undergraduate Student
Advanced to Research Associate at University of Iowa.
Davis Verhoeven
Undergraduate Student
Taylor Yoke
Undergraduate Student
William Young
Undergraduate Student
Advanced to Biomedical Sciences Graduate Program, Iowa State University.
Publications
Useful Links
Research Databases
Research Tools
University Resources
Affiliated Departments and Programs
Literature Resources
Safety Training and Information
Contact us